With the aim of assessing the conservation prioritization scores obtained through the gap analysis, a validation process was performed in collaboration with forty-four experts in the fields of botany, plant taxonomy, biodiversity and plant genetic resources conservation, and crop improvement, all of them selected by their experience with the wild relatives of crops (complete list of participating experts).
This process consisted of an online survey where the experts were asked to provide an evaluation score (or Expert priority score) representing the adequacy of existing CWR accessions in genebanks for each species, as well as the geographic and environmental gaps that may exist. The Expert priority score range is between 0 and 10, where 0 indicates that the expert considers that urgent efforts to increase the representativeness of the CWR taxon are required, and 10 that the CWR taxon is sufficiently represented in genebanks. An additional evaluation score (contextual expert priority score) was obtained to represent the experts’ knowledge in terms of usefulness of the taxon for plant breeding and threats in their natural habitats. The occurrence data points utilized as inputs of the gap analysis, prioritization score, the distribution maps, and collecting maps produced through the gap analysis were also subjected to the evaluation of experts. Finally, a combined index (Evaluation index) was estimated by applying a Multiple Factor Analysis (MFA) to summarize all the inputs received from experts, namely: expert priority score, comparable priority score and their assessment on the gap analysis products. The scale of this Evaluation index goes from 0 (disagreement) to 100 (agreement).
Here we display the results obtained for each crop.
Alfalfa (Medicago sativa)
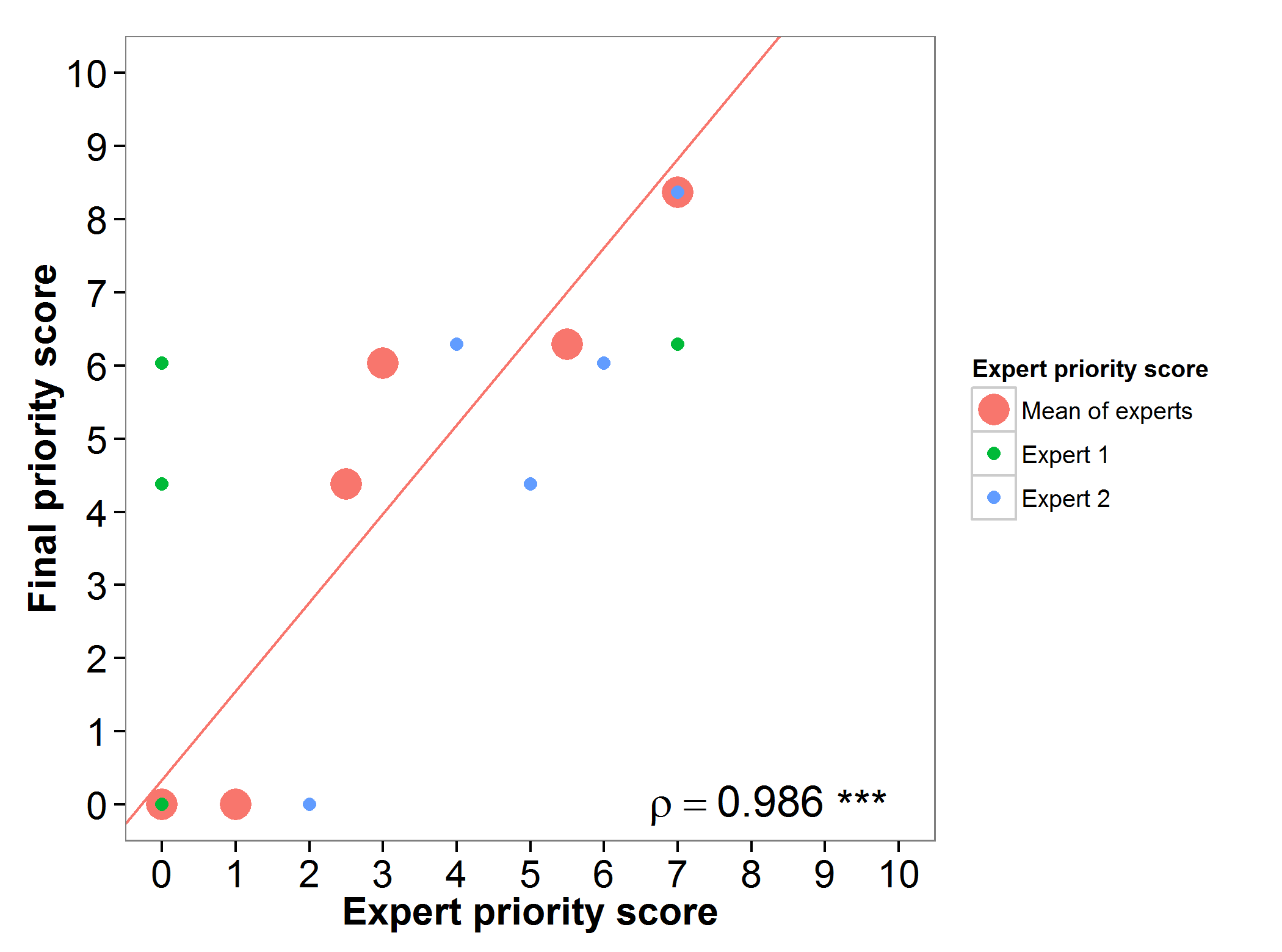
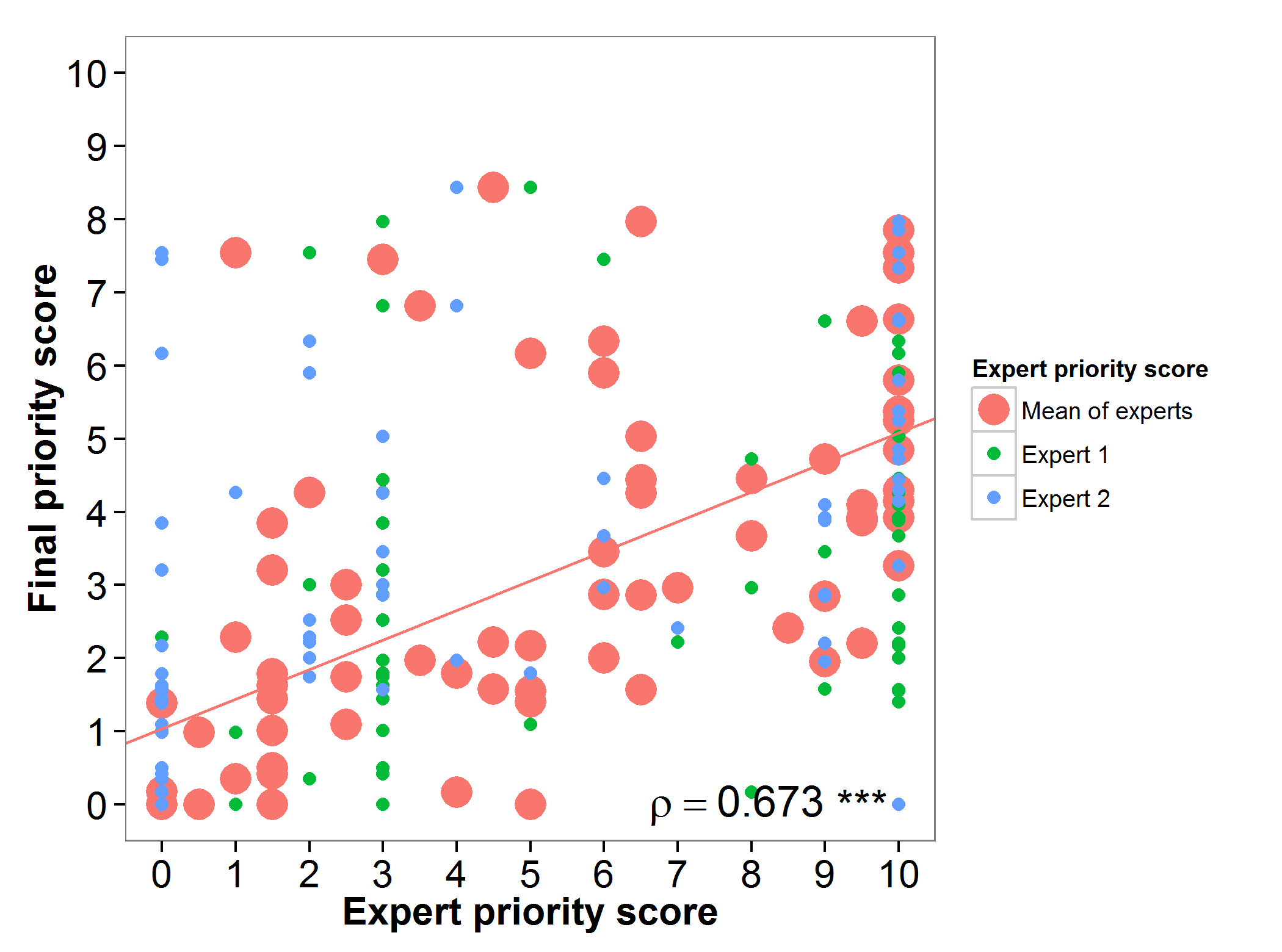
The final priority scores (FPS) obtained for alfalfa in the gap analysis were assessed by two experts with large experience in the conservation of this genepool. High agreement was found between the experts priority score and the gap analysis prioritization score (Figure 1).
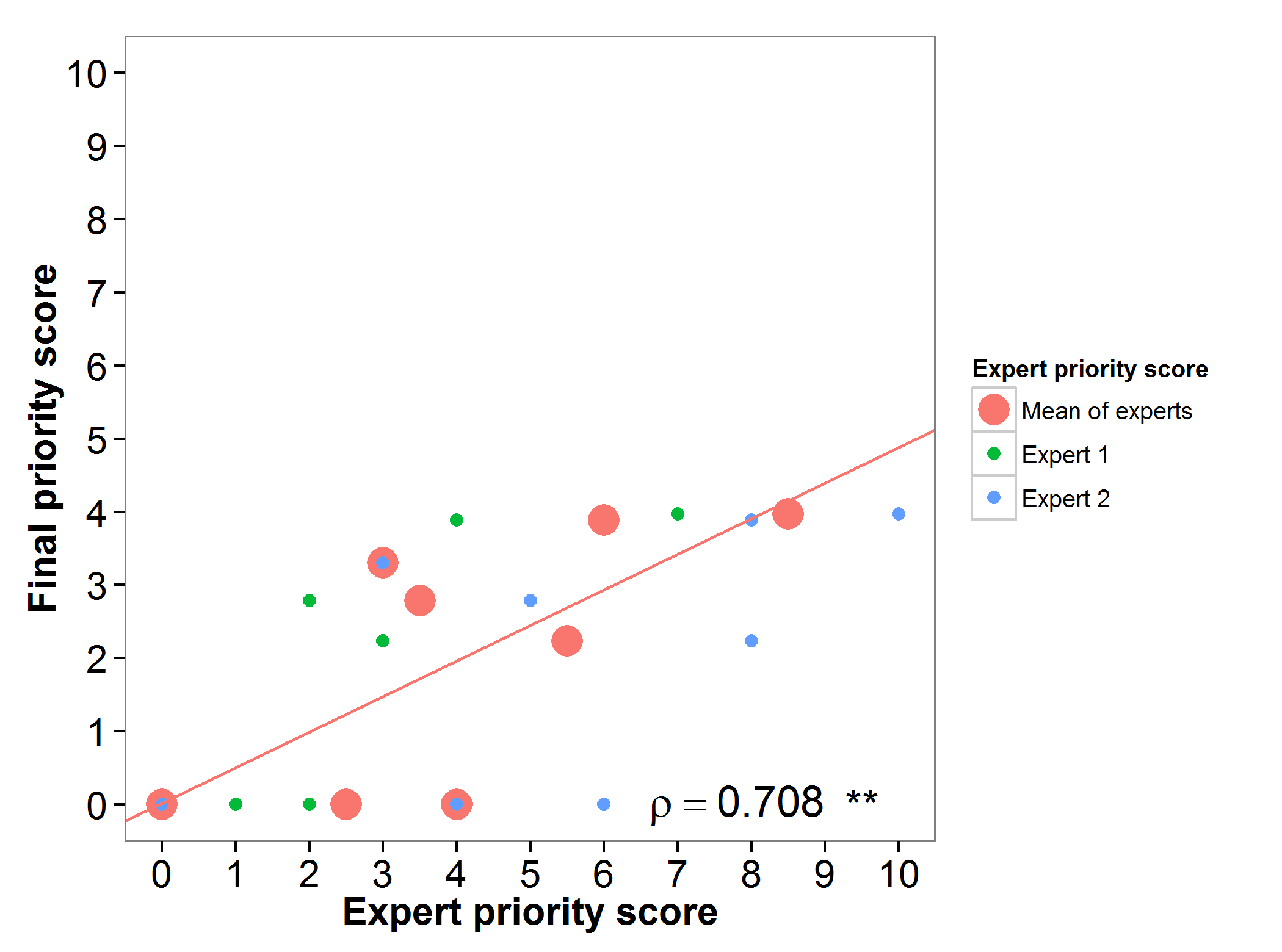
Figure 1. Relation between gap analysis prioritization results and comparable expert evaluation score per alfalfa wild relative.
Apples (Malus domestica)
The apple genepool was assessed by two experts. The evaluation index (Fig 2B) suggests an average agreement between experts and the gap analysis results, ten species (M. ombrophila, M. prunifolia, M. sikkimensis, M.hupehensis, M. yunnanensis, M. doumeri, M. mandshurica, M. orientalis, M. sieversii, M. sylvestris) obtained a medium to high evaluation index, while three species (M. fusca, M. pumila and M. zumi) obtained low evaluation index values.
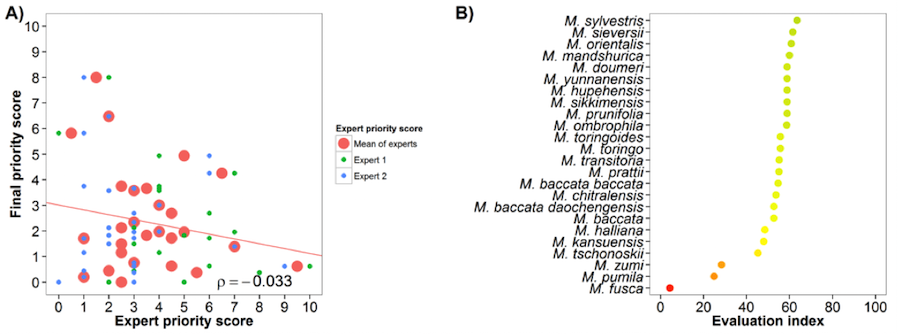
Fig 2. Expert evaluation agreement with gap analysis results for the apple genepool: A) relation between gap analysis results and expert evaluation scores. B) evaluation index per crop wild relative
Bambara groundnut (Vigna subterranea) and cowpea (Vigna unguiculata)
The wild relatives of bambara groundnut and cowpea were presented in a single survey to the expert, as both genepools belong to the genus Vigna.
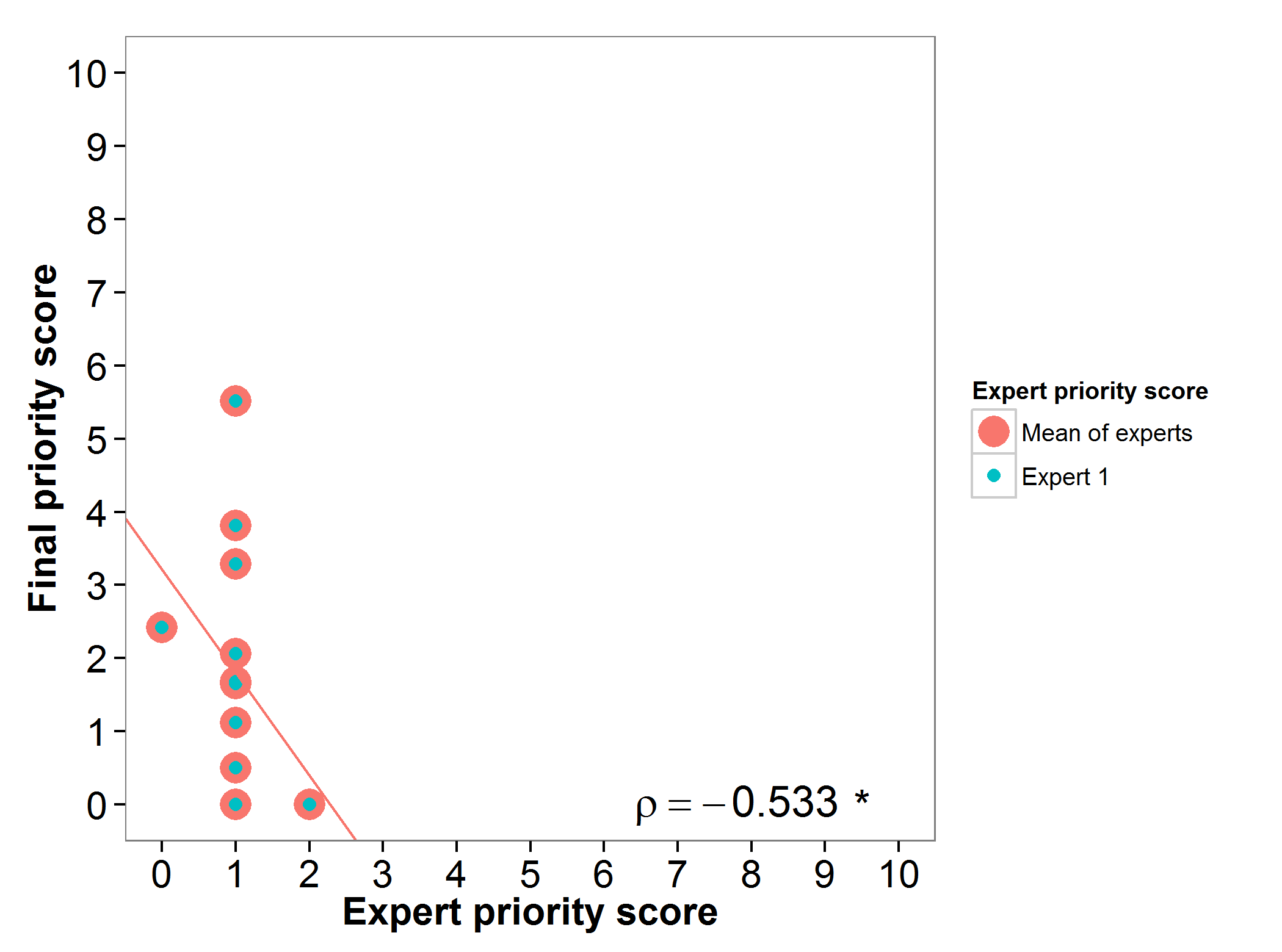
Fig 3. Relation between gap analysis prioritization results and comparable expert evaluation score perVigna wild relative
Bananas & plantains (Musa spp.)
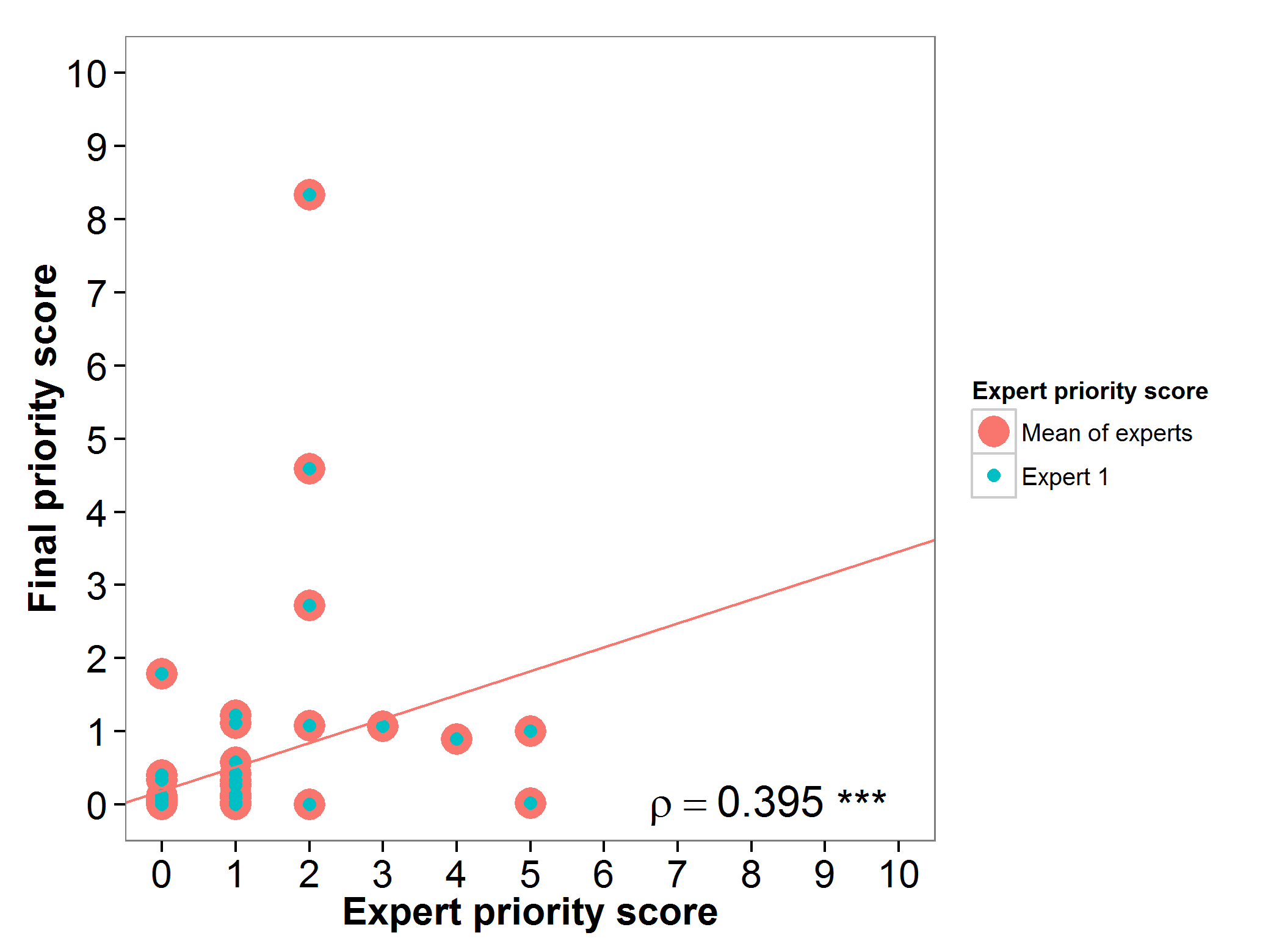
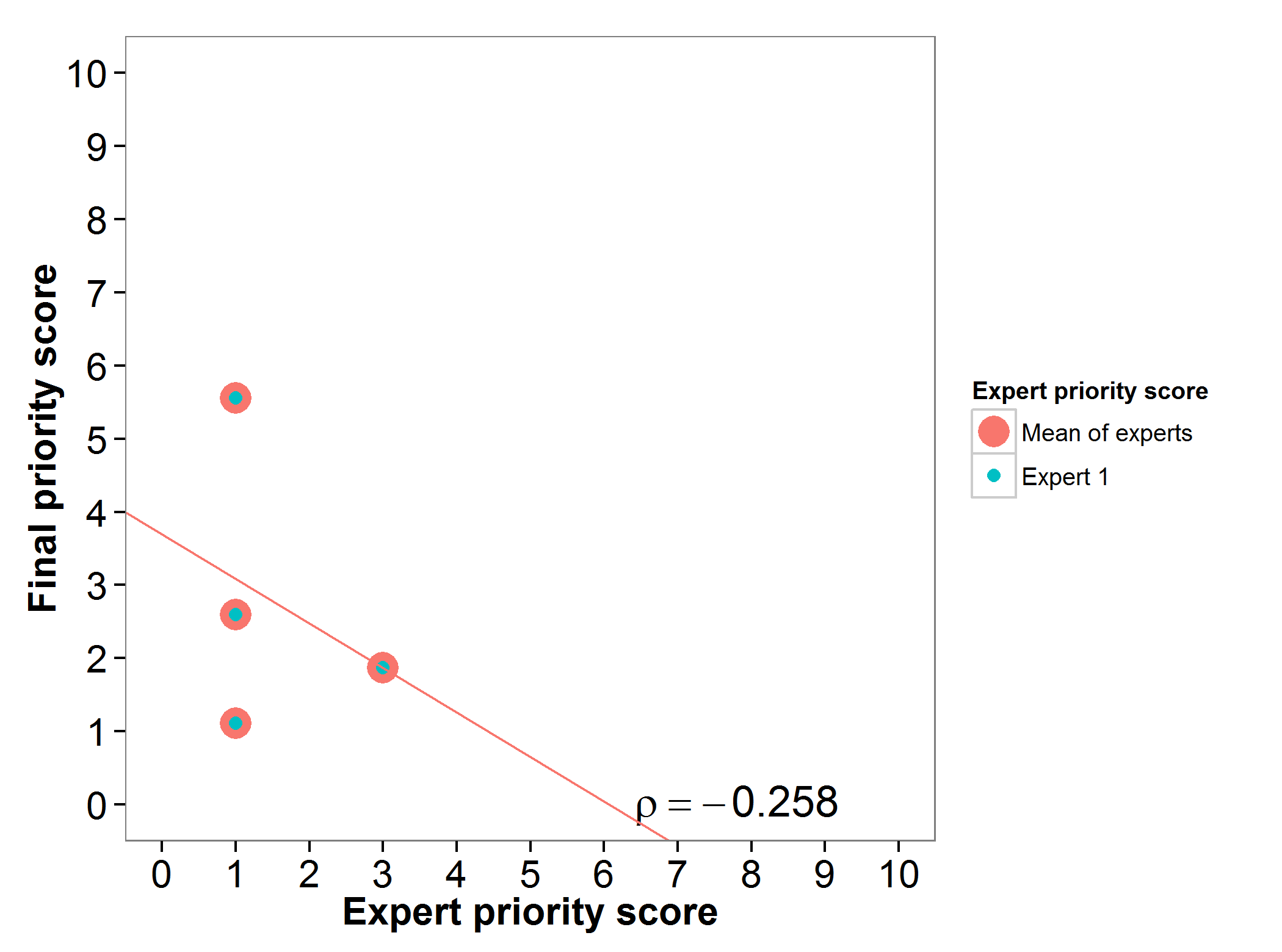
The genepool of bananas and plantains gap analysis prioritization scores were assessed by one expert, presenting an agreement between the experts prioritization score and the final prioritization score (Fig. 4).
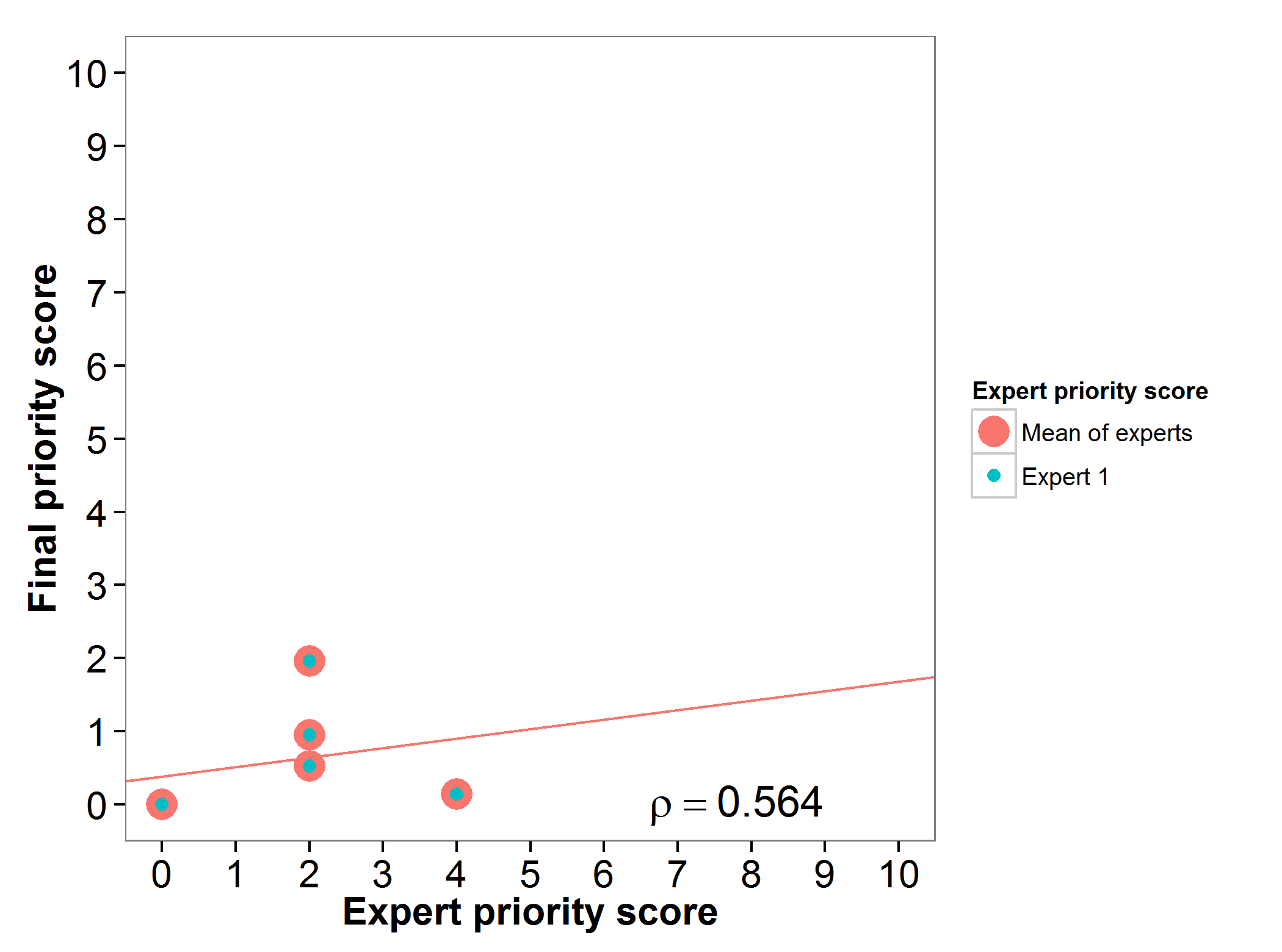
Fig 4. Relation between gap analysis prioritization results and comparable expert evaluation score per Musa wild relative
Barley (Hordeum vulgare)
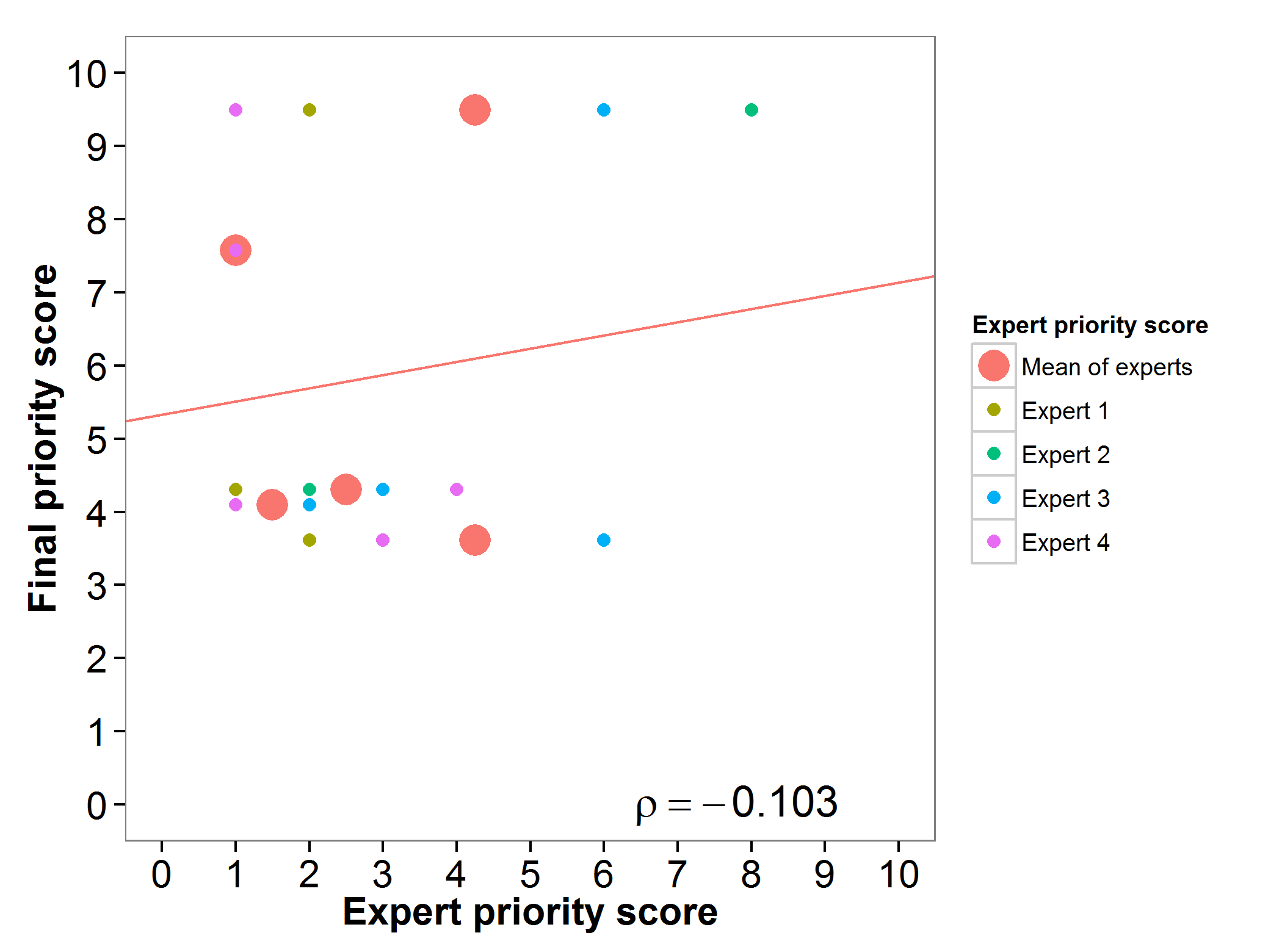
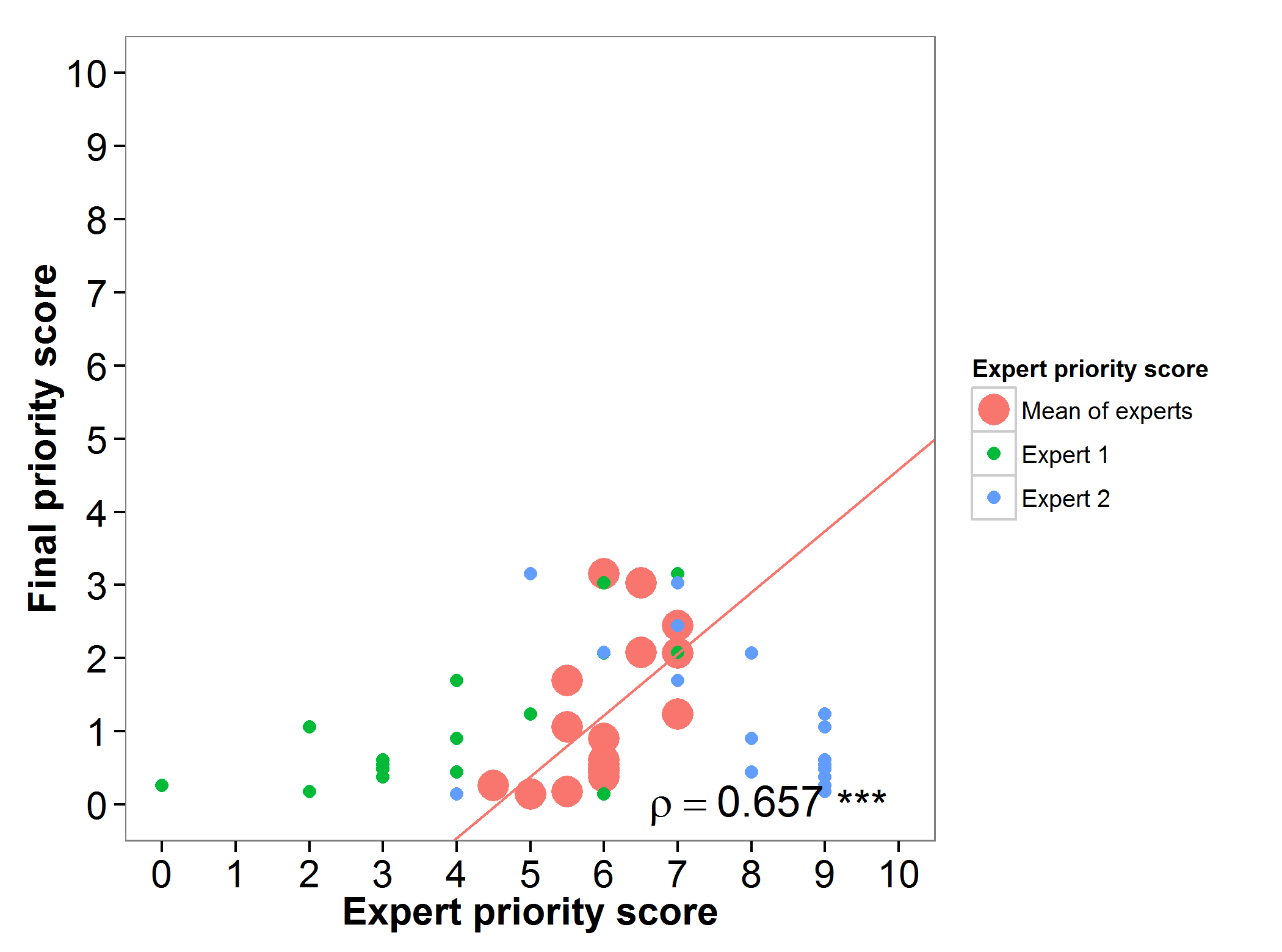
The final priority score obtained through the gap analysis for the wild relatives of barley were assessed by three experts. Large differences between the perceptions of the experts surveyed were evidenced for this genepool (Fig. 5).
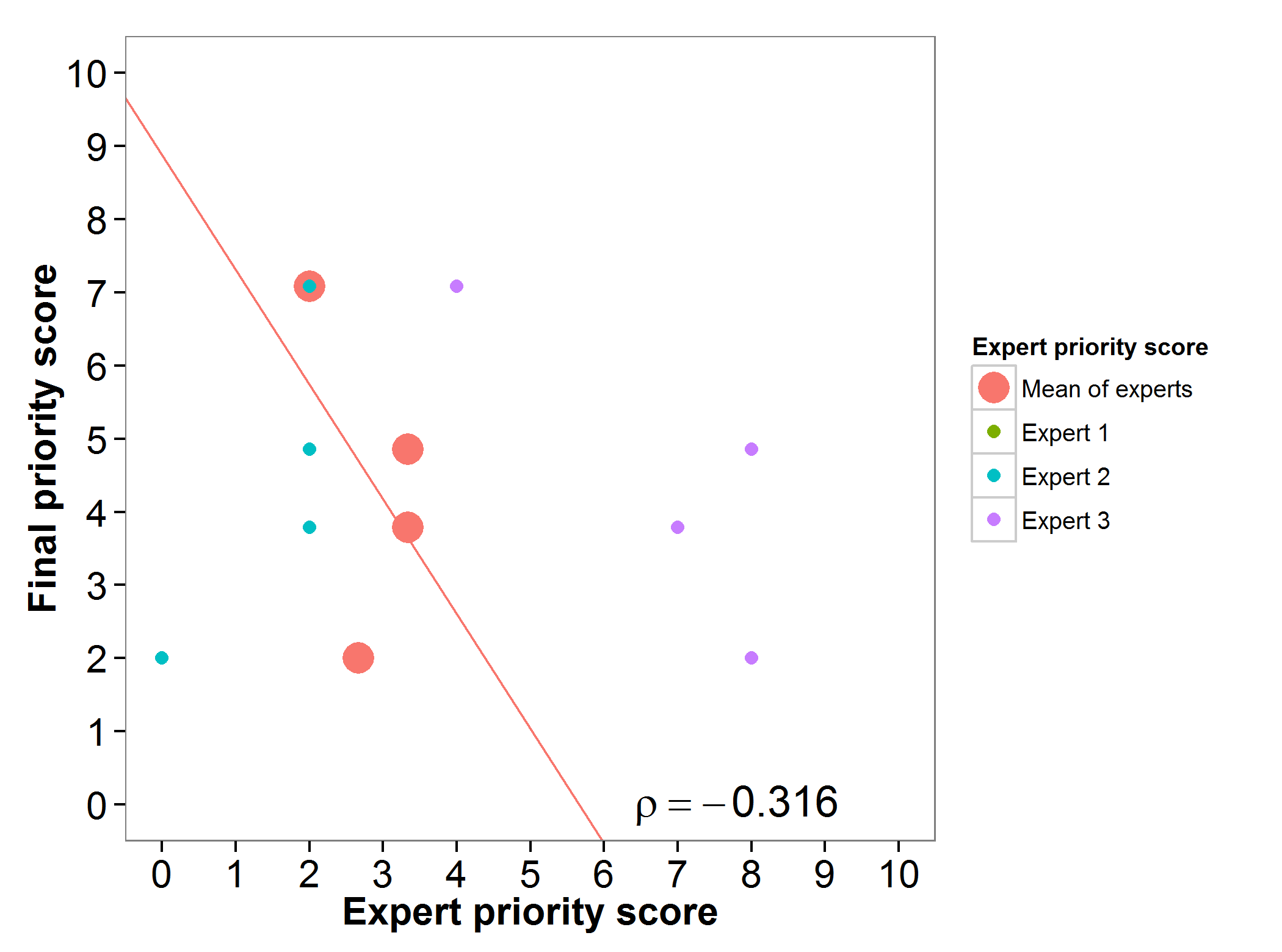
Fig 5. Relation between gap analysis prioritization results and comparable expert evaluation score per barley wild relative
Bean (Phaseolus vulgaris) and lima bean (Phaseolus lunatus)
The final priority score calculated through the gap analysis for Phaseolus wild relatives was evaluated by two experts, resulting in a high agreement between both scores (Fig. 6).
Carrots (Daucus carota)
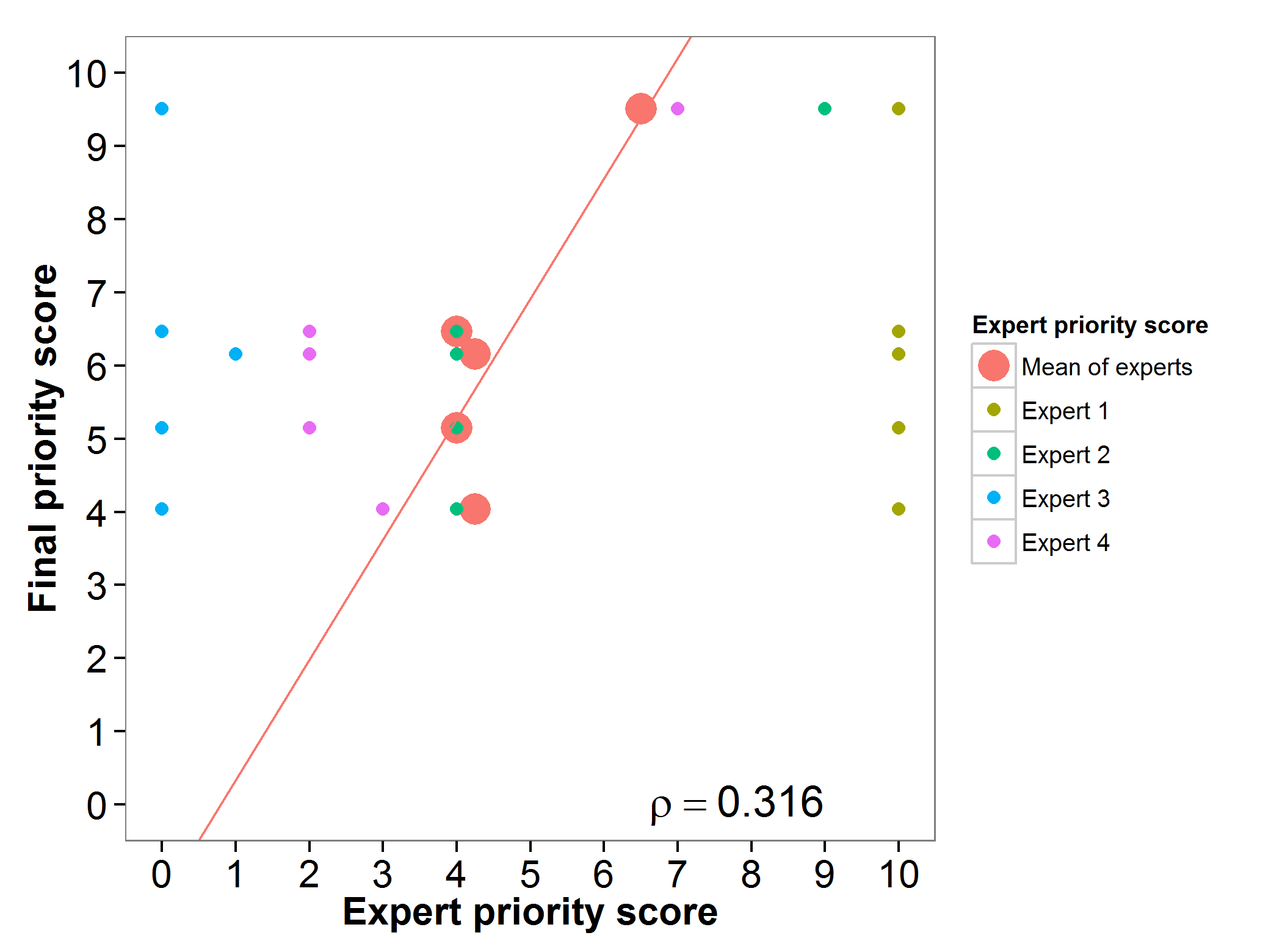
The carrot genepool gap analysis results were assessed by two experts, resulting in a high agreement among experts and the gap analysis final priority score (Fig. 7A). The evaluation index presents contrasting results, with two taxa having very low values, four having average values and four presenting high evaluation index values (Fig 7B).
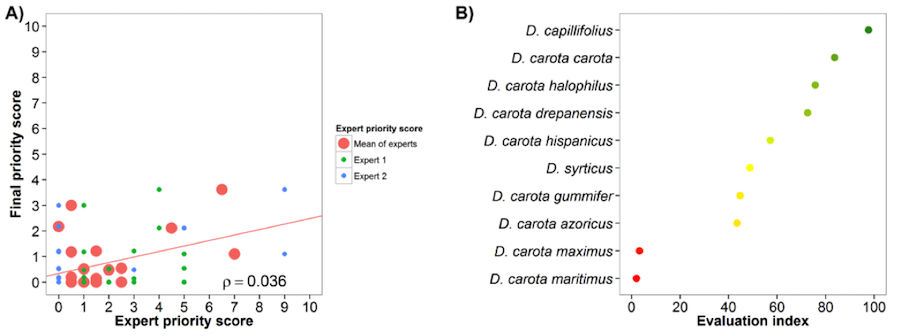
Fig. 7 Expert evaluation agreement with gap analysis results for the carrot genepool: A) relation between gap analysis results and expert evaluation scores. B) evaluation index per crop wild relative
Chickpea (Cicer arietinum)
The final priority scores of the wild relatives of chickpea were assessed by four experts (Fig. 8).
Eggplant (Solanum melongena)
The final priority scores of the wild relatives of eggplant presented a positive agreement with the expert assessment (Fig. 9).
Finger millet (Eleusine coracana)
The final priority scores of the wild relatives of finger millet were assessed by one expert. A positive agreement was found between the expert priority score and the gap analysis final priority score (Fig. 10).
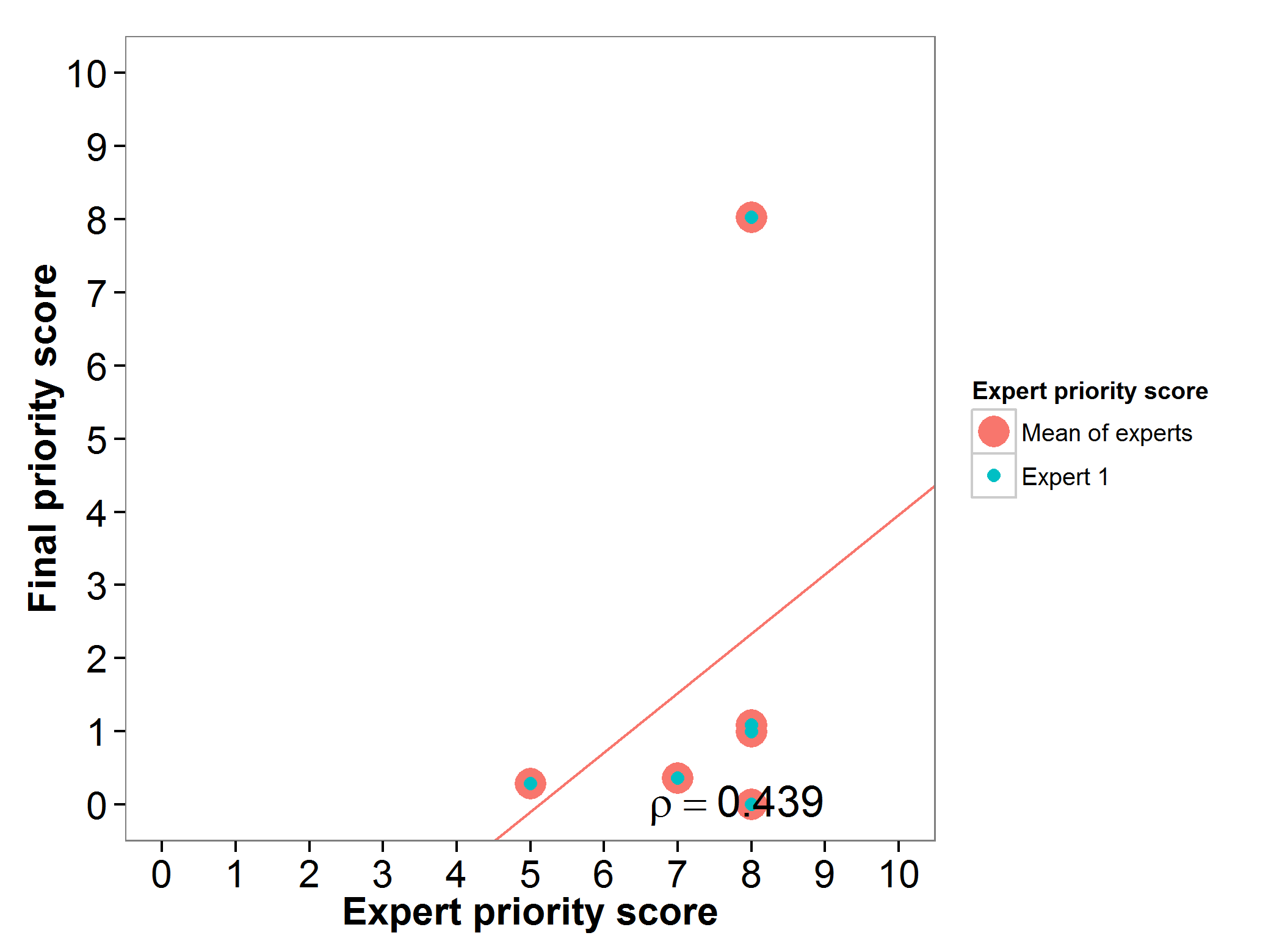
Fig 10. Relation between final priority score and expert priority score per finger millet wild relative
Grasspea (Lathyrus sativus)
The final priority scores of the grasspea genepool were assessed by four experts. A high agreement was obtained as reflected in Fig. 11.
Lentil (Lens culinaris)
Oats (Avena sativa)
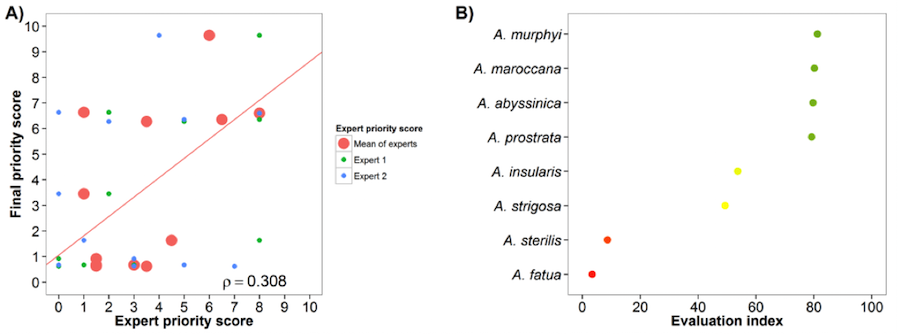
Fig. 13 Expert evaluation agreement with gap analysis results for the oat genepool: A) relation between gap analysis results and expert evaluation scores. B) evaluation index per crop wild relative
Peas (Pisum sativum)
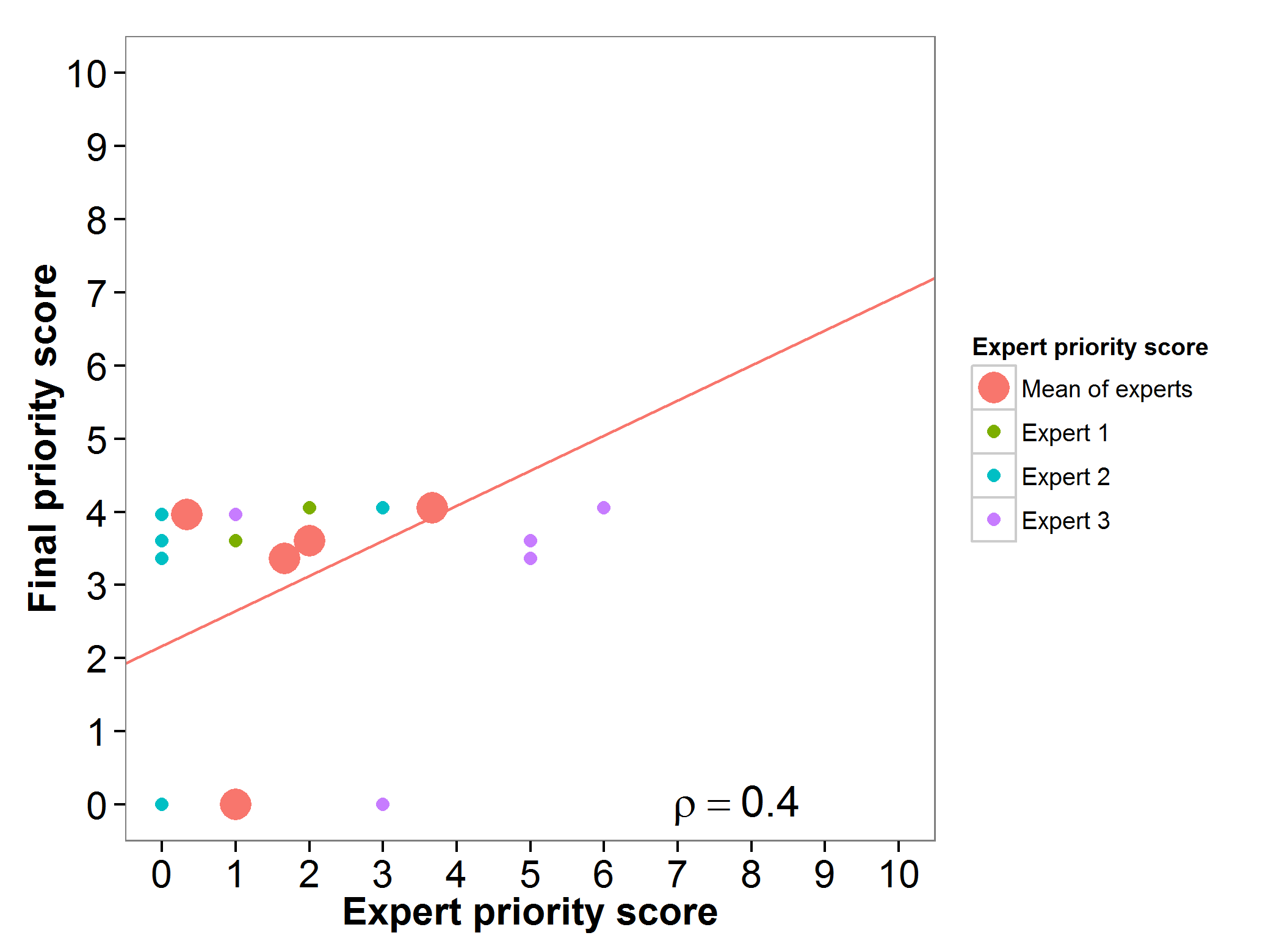
Fig 14. Relation between final priority score and expert priority score per Pisum sativum wild relative
Pearl millet (Pennisetum glaucum)
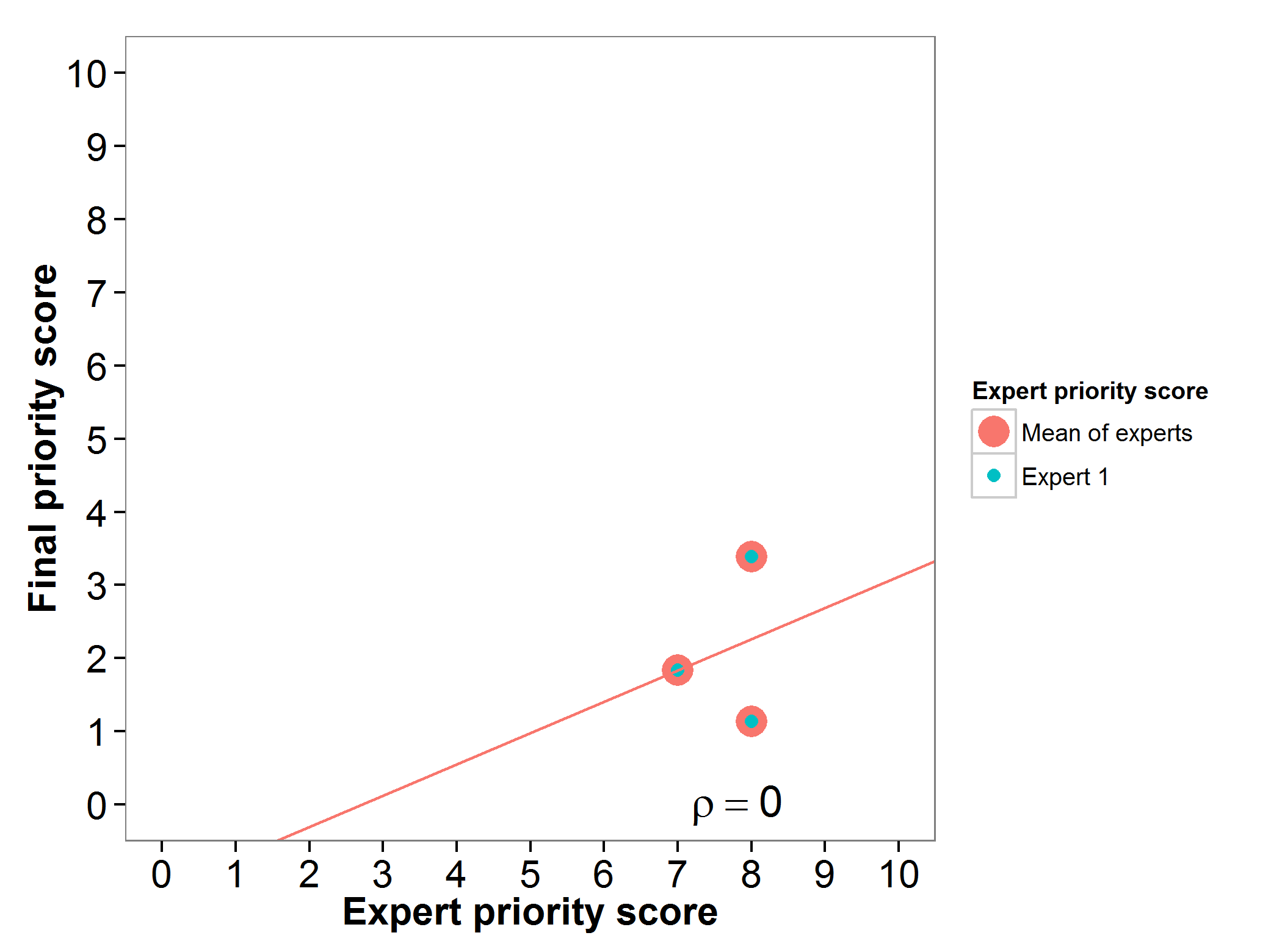
Fig 15. Relation between final priority score and expert priority score per pearl millet wild relative
Pigeonpea (Cajanus cajan)
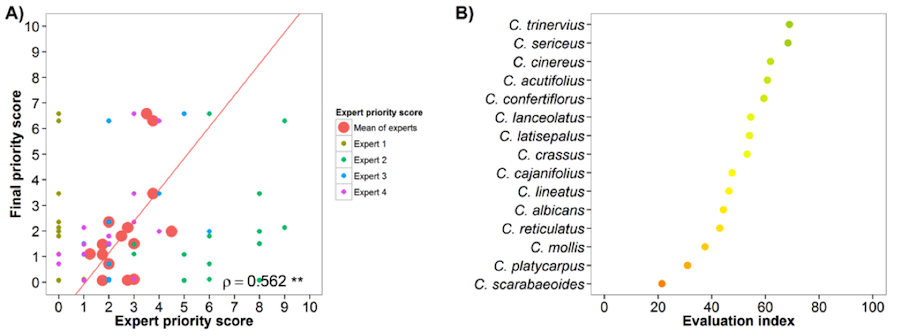
Fig. 16 Expert evaluation agreement with gap analysis results for the pigeonpea genepool: A) relation between gap analysis results and expert evaluation scores. B) evaluation index per crop wild relative
Potato (Solanum tuberosum)
Rice (Oryza sativa) and African rice (Oryza glaberrima)
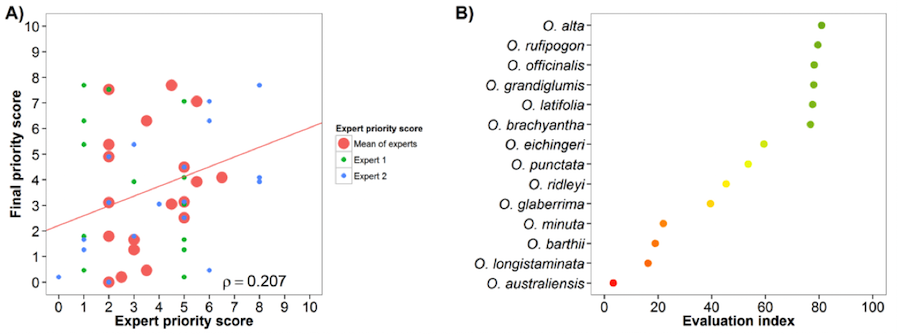
Fig. 18 Expert evaluation agreement with gap analysis results for the rice genepools: A) relation between gap analysis results and expert evaluation scores. B) evaluation index per crop wild relative
Rye (Secale cereale)
Sorghum (Sorghum bicolor)
Sunflower (Helianthus annuus)
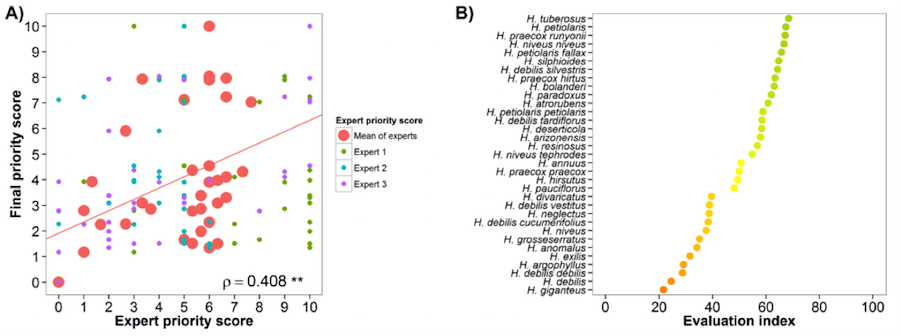
Fig. 21 Expert evaluation agreement with gap analysis results for the sunflower genepool: A) relation between gap analysis results and expert evaluation scores. B) evaluation index per crop wild relative
Sweetpotato (Ipomoea batatas)
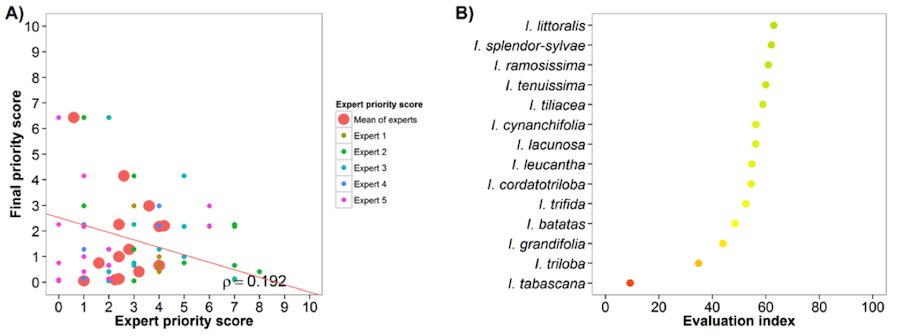
Fig. 22 Expert evaluation agreement with gap analysis results for the sweetpotato genepool: A) relation between gap analysis results and expert evaluation scores. B) evaluation index per crop wild relative
Wheat (Triticum sativum)
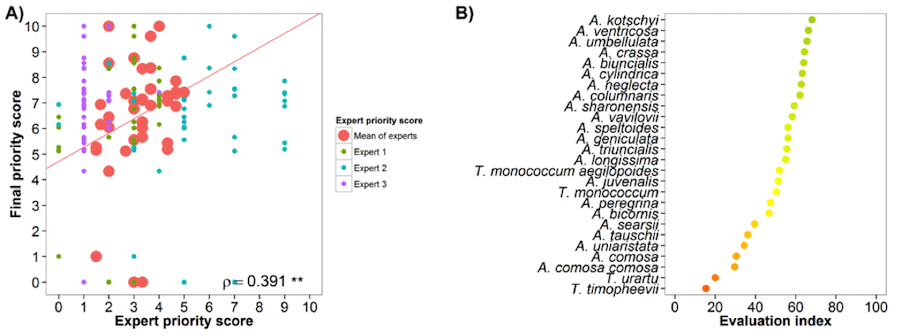
Fig. 23 Expert evaluation agreement with gap analysis results for the wheat genepool: A) relation between gap analysis results and expert evaluation scores. B) evaluation index per crop wild relative
Collecting and Conservation
A quarter of our plant diversity is threatened globally due to industrial agriculture and forestry practices, urbanisation, over-collection, pollution, land use changes, the spread of invasive alien species and climate change. CWR are also subject to these threats. There is an urgent need to collect CWR, an important building block for crop breeding, to ensure all options for the future are maintained.
The scale of the collecting need is too large to be completed by one organisation alone, thus a concerted and prioritised effort is needed. This is recognised in the Global Strategy for Plant Conservation where target 9 includes the conservation of CWR. The gap analyses available on this website present one powerful form of prioritisation. The results here can be used to target and co-ordinate collecting efforts at local, national, regional and global levels.
Ex situ CWR collections are critical for conservation, and form a vital source of plant genetic resources for breeding. This significance for breeding can only be realised if collections are available to breeders and other researchers. Stumbling blocks to availability include small seed collections, insufficient resources to carry out distribution, lack of information, and political decisions. Benefit sharing is a major aspect to consider and several mechanisms are in place to address this. The Convention on Biological Diversity (CBD) has introduced access and benefit sharing requirements and recently the Nagoya Protocol to the CBD has further specified the requirements. For a determined list of crops and their CWR, the International Treaty on Plant Genetic Resources for Food and Agriculture (ITPGRFA) has established a specialized access and benefit-sharing regime. The project operates its collecting phase in the framework of the ITPGRFA, in full respect of applicable domestic regulatory requirements.
Collecting partners
- Cyprus: The Agricultural Research Institute of the Ministry of Agriculture, Natural Resources and Environment and collaborator ICARDA.
- Georgia: The Institute of Botany and the National Botanical Garden of Georgia.
- Italy: Department of Earth and Environmental Sciences of the University of Pavia and collaborators; University of Cosenza, Foreste Casentinesi Mount Falterona and Campigna National Park, State Forestry Corps, Command of Punta Marina (Ravenna, Emilia-Romagna), Province of Ravenna.
- Kenya: Kenya Agricultural and Livestock Research Organisation and collaborators; National Museums of Kenya, Kenya Wildlife Services, Kenya Forestry Service and Kenya Forestry Research Institute.
- Portugal: Museu Nacional de História Natural e da Ciência and collaborators; Banco Portugues de Germplasma Vegetal, ISOPlexis Banco de Germoplasma (Universidade da Madeira) and Jardim Botanico do Faial – Banco de Sememtes dos Acores.
- Vietnam: National Plant Genetic Resources Institute and collaborators; The Northern Mountainous Agriculture and Forestry Institute, Vietnamese Academy for Agriculture Sciences and Cuulong Delta Rice Research Institute.