Category : Feature Stories
Published : August 13, 2018 - 1:01 PM
Michael Major, Crop Trust
Pre-breeders generate a lot of data. A LOT of data. They can make thousands of crosses between wild and domesticated species of food crops and evaluate those thousands of crosses under various conditions, in different climates and countries. Then they’ll make backcrosses and evaluate those crosses. Collecting and managing the data is hard work but analyzing it is an even bigger challenge – but one that must be addressed if pre-breeding is going to contribute to the development of studier ‘climate-proof’ crops.
The Crop Wild Relatives (CWR) Project coordinated by the Crop Trust is managing pre-breeding projects on 19 crops. “These projects are bringing back to our most important crops the many useful traits that their wild cousins still have in the genetic make-up, but the crops themselves have left behind. By crossing and backcrossing these plants, our partners are generating complex data in huge quantities,” said Hannes Dempewolf, Head of Global Initiatives at the Crop Trust. “For example, the sunflower pre-breeding project resulted in 545,000 molecular markers. This kind of data is an amazing contribution to the breeding community. We wanted to make sure it is publicly available, so we could maximize the number of breeders around the world that use it, and the plants themselves, in their improvement programs.”
Pre-breeding data available to everyone
The Crop Trust teamed up with the James Hutton Institute in Invergowrie, Scotland to ensure the CWR project’s pre-breeding data is available in a format that allows breeders and scientists to view and analyze the data as easily as possible. Hutton has been developing software known as Germinate which is specifically tailored to handle complex data from the use of plant genetic resources collections.
“We wanted to create a tool so that researchers and breeders can share data about different crops on a customizable, yet common, platform,” said Paul Shaw, a research leader in Information Systems at Hutton. “So we developed Germinate, which is a database system that can be used to view and select plant genetic resources data and then analyze it using various visualization tools.”
It became apparent that Germinate 3 – the latest version of Germinate – would be a perfect fit for the CWR project’s pre-breeding data. “We began looking at Germinate 3 as a platform for us to share our pre-breeding data with the world back in 2014,” said Hannes. “We wanted an easy-to-use tool that allows users to drill down through our partners’ massive datasets and make decisions which would help in their breeding or research activities. We felt the James Hutton Institute was ideally suited to lead this effort due to their experience in handling such data.”
These days new technologies are being developed which are making it much easier and cheaper to generate huge amounts of phenotypic and genotypic information. But storing data is just a start. Users need to be able to find the data they require, so presenting the data in a user-friendly and intuitive interface is equally important.
“Germinate 3 fills a role not offered by other plant genetic resources software platforms,” said Paul. “In short, it is capable of integrating both genotypic and phenotypic data with passport data.”
Additionally, Hutton has developed versatile graphical search functions on Germinate 3. “These allow users to identify groups of samples that meet selected passport, molecular or phenotypic criteria,” said Sebastian Raubach, Hutton’s Bioinformatics Software Developer who has worked extensively on the development of Germinate 3. “Once users have identified the data they are interested in, they can download it in a variety of formats.”
But the real power of Germinate 3 lies in its ability to integrate with a range of external data visualization software. These programs allow breeders to review large datasets in easily digestible graphics.
“We built functions into Germinate 3 which allow plant breeders and other scientists to export the data and then import it into these visualization programs,” said Paul. “That means users can perform complex analyses of the data outside of Germinate 3 and generate data-rich graphics.”
Hutton has also developed several external programs which provide user-friendly analysis of large datasets. Helium can help breeders determine the “genealogy” of a plant line. Flapjack helps users compare lines, markers and chromosomes by visually displaying similarities. CurlyWhirly can help users find patterns and outliers in the data.
Thus far, the Hutton team have created Germinate 3 pilot platforms for rice and sunflower using the CWR project’s pre-breeding data. Whereas the species may differ, the approach taken ensures that the tools are compatible, and developments can benefit all crops. In other words, work that Hutton has done on the rice database will benefit the Crop Wild Relatives’ durum wheat project, and all the others as well.

Germinate 3 developers Sebastian Raubach (left) and Paul Shaw (right) presented Germinate 3 at the Royal Highland Show in June 2018.
Going Live
Pre-breeding data from the below crops will be made available on Germinate 3
| Alfalfa | Barley |
| Chickpea | Cowpea |
| Durum wheat | Eggplant |
| Finger millet | Grass pea |
| Lentil | Pearl millet |
| Pigeonpea | Rice |
| Sorghum | Sunflower |
“In the following months, we will be developing and deploying a series of web portals using Germinate 3 to support access to data from 14 of our CWR pre-breeding projects,” said Benjamin Kilian, Plant Genetic Resource Specialist at the Crop Trust. “We are now ready to launch the portal for the eggplant pre-breeding project.” The CWR Eggplant Database lists data on nearly 1,000 eggplant samples and more than 1,500 molecular markers.
“The Eggplant Database will give us an opportunity to receive some user feedback concerning Germinate 3,” said Benjamin. “This will help Hutton further improve on the product, as we continue releasing the data from our partners’ other pre-breeding projects.”
The CWR pre-breeding projects will continue to generate important data that will help plant breeders improve many of our food crops, making them more resilient to climate change. With Germinate 3, plant genetic scientists and breeders can rest assured that this data will long remain readily available on a versatile and powerful platform.
###
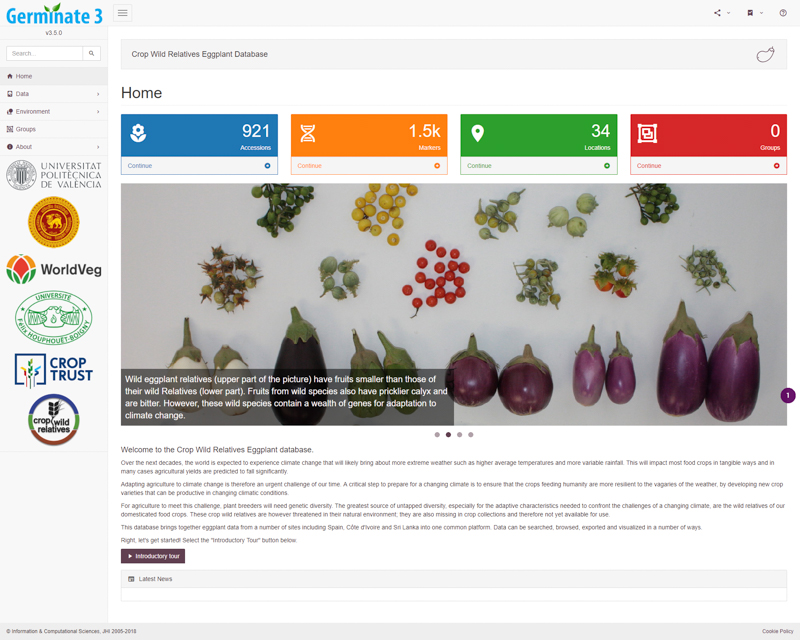
Germinate provides a standard and common interface to genetic resources collections. Each Germinate database looks and feels the same, but color and branding are used to differentiate instances. This consistent presentation allows users to become familiar with Germinate no matter which species they are working with and visualization and data query tools are familiar across species.
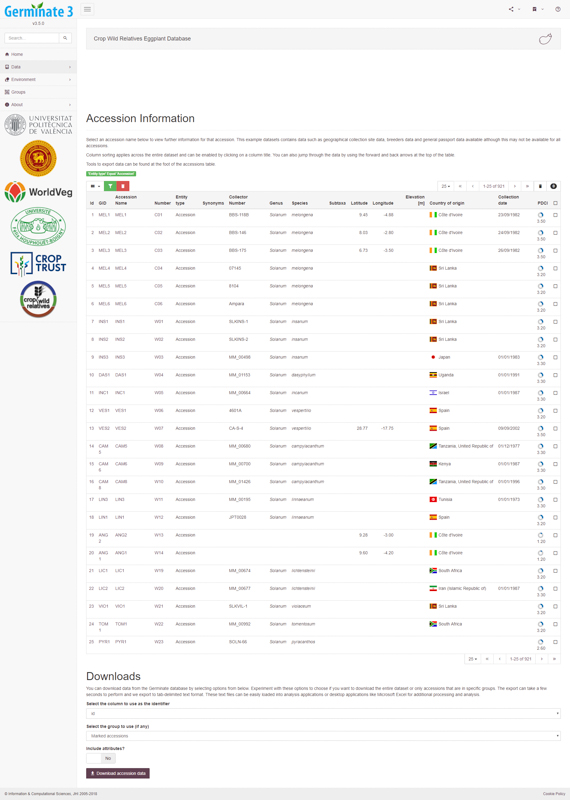
Germinate users can select germplasm from lists based on a number of categories such as country of collection, collecting altitude, species name and collecting dates. Each of these factors can be layered to provide a powerful and intuitive search system. Once users find germplasm which matches their requirements they can add it to a ‘shopping cart’ which allows groups of germplasm to be collected and stored within Germinate. These can then be used to export the appropriate genotypic and phenotypic data from datasets stored within Germinate, for use in visualization tools.
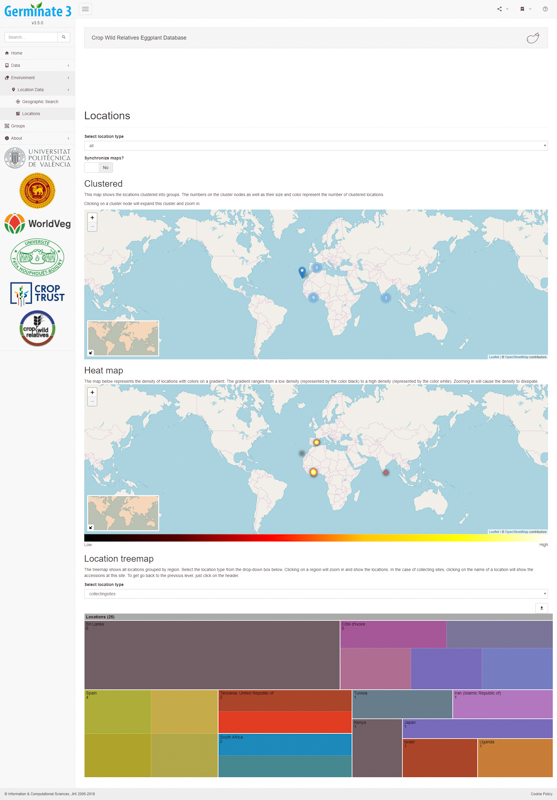
Data overview statistics give users an understanding of the geographical distribution of data contained within Germinate. The software uses heat maps to show where germplasm was collected and uses novel information visualization techniques such as tree maps to give an indication of the number of accessions from different countries. A user can use these sorts of maps to choose germplasm from specific geographic regions and create a Germinate group to be used later in data export.

Phenotypic field trials data stored in Germinate can be queried in a number of ways. In this example, scatter plots show all possible pairwise comparisons among five different traits. These charts are useful for showing the relationship between phenotypes and can highlight outliers which may be of interest to plant breeders or an indication of problematic data which requires closer inspection. A Germinate user can select germplasm based on the selection of points in these charts using lasso tools. Selection of a data point in one scatter plot will highlight the corresponding data in other charts, making data exploration interactive in real time.
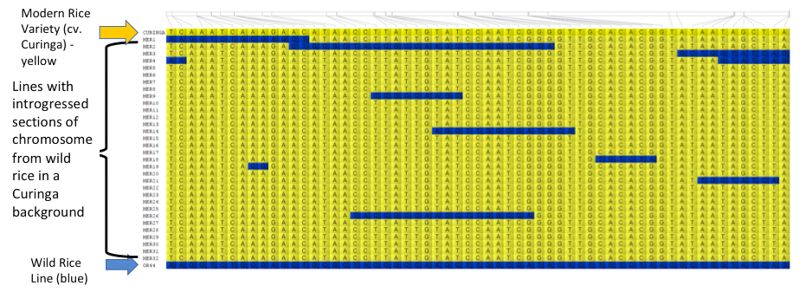
Genotypic data is also stored in Germinate and can be exported into tools such as Flapjack (https://ics.hutton.ac.uk/flapjack), a graphical genotype viewer. Users can move from Germinate to Flapjack then back again to Germinate seamlessly in order to get additional information on plant lines or markers. Data can also be exported to other visualization tools, such as Helium and CurlyWhirly where appropriate, and these tools are freely available from https://ics.hutton.ac.uk. This Flapjack image shows the genotypes of rice pre-breeding lines based on 61 molecular markers located on chromosome 1. The yellow cells show modern rice background while the blue cells show wild rice genomic fragments.
All material collected under the Crop Wild Relatives project is shared under the terms of the Standard Material Transfer Agreement (SMTA) within the framework of the multi-lateral system of the International Treaty for Plant Genetic Resources for Food and Agriculture (ITPGRFA).
All data is publicly available and shared under a Creative Commons license CC 4.0. A short demonstration video of how the license is implemented can be found here. Furthermore, all users of the database are encouraged to use a system of unique identifiers when referencing germplasm, called DOIs, as implemented by the Global Information System under the ITPGRFA.
